
DEPArray™ NxT 單細胞分離篩選系統
隨著 DEPArray™技術的發展,基因的異質性研究可達單顆細胞層級。可探索每顆細胞基因中隱藏的故事,例如整倍體的基因變異。DEPArray™技術可依細胞表型分選,以及運用不同參數組合與影像系統的雙重確認,可確保分選出純度 100% 的標的細胞,採用高純度細胞故可觀察出等位基因表現。
分類:DEPArray 單細胞分離篩選系統 > 研究儀器
技術原理
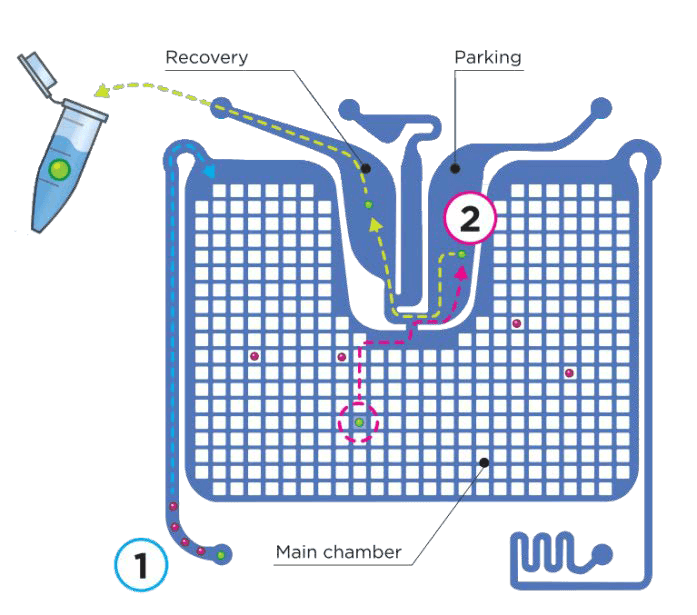
DEPArray™獨特技術建立在晶片內形成介電場,使細胞在電場中懸浮,並排列整齊。
富集過後的細胞,經過螢光染色後可注入晶片。晶片置入 DEPArray™ NxT 機台後,細胞會自動進入主要晶片區,進行分選。
晶片中的電場活化後形成電籠包裹細胞懸浮,儀器有 6 個螢光通道可供選擇,搭配影像系統可用來分析細胞表現,篩選出目標細胞。高質量影像系統搭配電極控制電籠移動,精確地進行細胞分選和回收。
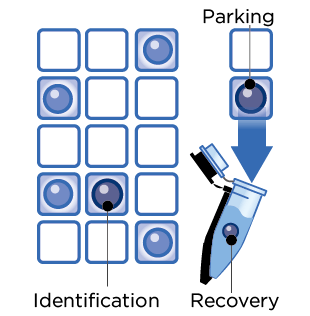
每一顆被選取的細胞會自動移動到「暫存區」,一旦完成,便可將細胞單顆 / 多顆單管回收到微管,進行後續分子層級的分析。
系統介紹
- 桌上型系統平台,節省空間
- 精確分離目標細胞,純度 100%,獲得後續二代測序準確信息
- 多樣化樣品上樣,活細胞,固定細胞皆可
- 自選標定用抗體種類

- 全新晶片設計,可回收高達 96 管單顆細胞
- 電籠包裹細胞移動,對細胞傷害性小,更適合活細胞分選
- 已設定標準化分選程序,統一篩選條件,目前應用在 CTC、FFPE、法醫檢體方面
- 可根據實驗設定,自定義篩選條件
- 高質量成像,圖像式操作界面,高度自動化
- 一部電腦可控制多台儀器,擴充方便
挑選與收集細胞
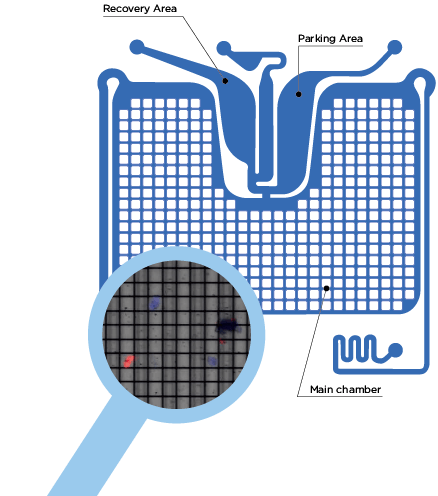
DEPArray™晶片
- 注入樣品,擷取細胞影像
- 移動目標細胞到暫存區
- 回收目標細胞
高質量成像
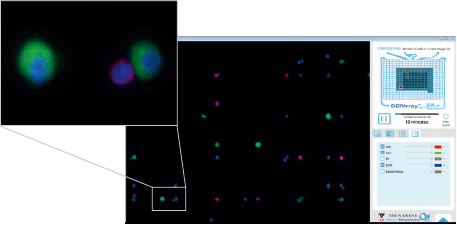
- 每個細胞都拍攝高質量影像
- 同時掃描,同時分析影像,自動將不同螢光影像選圖
- 每個細胞分析後給予獨立 ID,並記錄相關參數
人性化設計
- 操作者、樣品、芯片、回收盤,皆可使用掃描條形碼方式輸入,節省時間
- 自動化整理實驗報告,以 PDF 檔輸出
- CTC、FFPE、法醫檢體方面,默認篩選模組,統一分選標準。
- 觸控面板,圖像化設計,方便操作

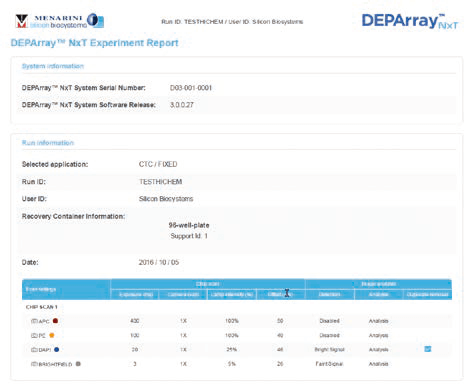
CTC 循環腫瘤細胞應用
DEPArray™ NxT 於 CTC 應用上的優勢
腫瘤是基因高度異質性的疾病,可能由基因突變引起,不論是遺傳或後天都有可能。大部分腫瘤都有特殊的分子改變,了解分子層級的變化有助於診斷和藥物開發。
- 捕獲單顆腫瘤細胞,甚至是循環腫瘤細胞 (CTC)
- 確認有上皮 – 間質轉化現象的相關亞型,或發掘新的亞群
- 分辨腫瘤亞型特徵
- 各種腫瘤細胞來源皆可分析:石蠟包埋組織,血液,細針活穿樣品等。
隨著 DEPArray™技術的發展,基因的異質性研究可達單顆細胞層級。可探索每顆細胞基因中隱藏的故事,例如整倍體的基因變異。DEPArray™技術可依細胞表型分選,以及運用不同參數組合與影像系統的雙重確認,可確保分選出純度 100% 的標的細胞,採用高純度細胞故可觀察出等位基因表現。
完整流程規劃
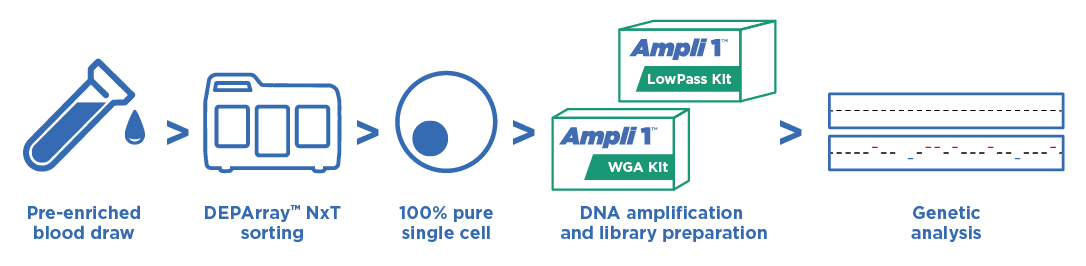
後續分析
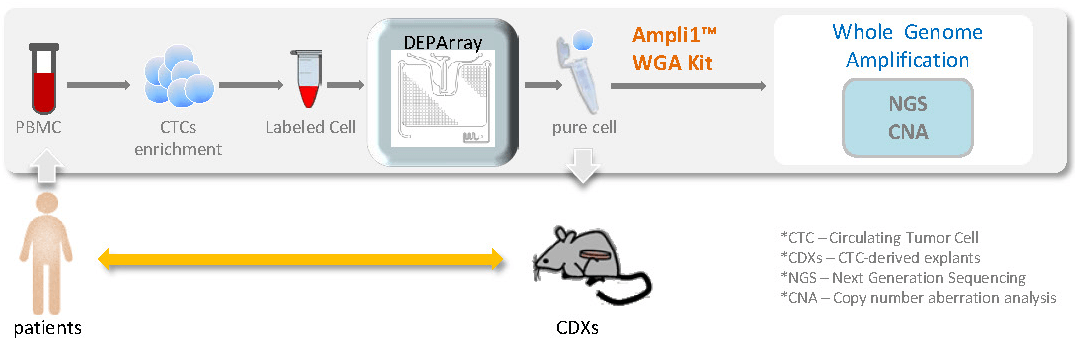
在腫瘤研究上,從外周血液中經由富集和篩選可純化出循環腫瘤細胞 (CTC),透過全基因組放大,有足夠的 DNA 產物進行後續不同的分子層級分析。單顆細胞的研究有助於解開腫瘤細胞間的異質性。全面性了解腫瘤轉移的機制。
分離出的單顆細胞,可使用 Ampli1™ WGA Kit 進行全基因組放大,再以 Ampli1™ LowPass Kit 處理後,可以進行二代定序,結果可進行 CNV 分析,可以明顯分辨出腫瘤細胞基因突變位點。
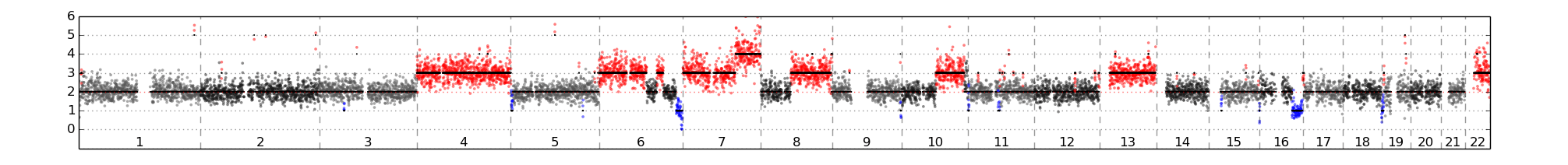
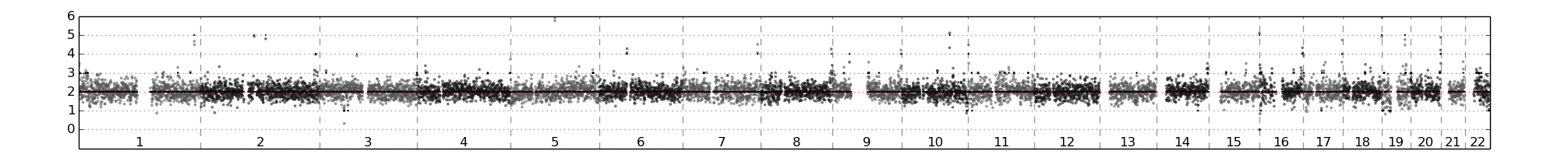
CTC 循環腫瘤細胞實際應用
2014 Nature medicine
Tumorigenicity and genetic profiling of circulating tumor cells in small-cell lung cancer
研究者介紹
Prof. Caroline Dive 是英國曼徹斯特大學藥理學教授,也是曼徹斯特癌症研究中心的傑出研究員 (Manchester Cancer Research Centre),她與她的研究團隊致力於以非侵入性的方式,透過偵測血液中的循環腫瘤細胞 (Circulating tumor cells),歸納出可以實際應用於疾病診斷以及追踪治療效果的數據庫,達到轉譯醫學的目的。
實驗設計
論文摘要
小細胞肺癌 (SCLC) 在肺癌中佔 15~20%,早期就轉移,預後差,多數病人無法以手術方式去除腫瘤。Prof. Caroline Dive 將病人周邊血液中的循環腫瘤細胞 (CTC) 分離出來,成功在免疫不全的老鼠建立疾病模式 (CTC-derived explants,CDXs),與病人相似,可藉由動物模式測試藥物,選取最適合藥物進行治療,邁向個人化醫療。文中並比較 CTC 與 CDXs 在基因表現上高度相關,CTC 有益於監控病程,CDXs 有助於研究抗藥性的作用機制。
2016 Nature medicine
Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemosensitive and chemorefractory small-cell lung cancer
論文摘要
Prof. Caroline Dive 利用已經建立的實驗流程,以 DEPArray 分離出已知化療敏感或化療不敏感的病患 CTC,進行基因檢測,找出 16 個基因組可以預測化療反應。之後在病人與老鼠模式上都可以成功預測,辨識正確率達 83.3%。
FFPE 組織蠟塊應用
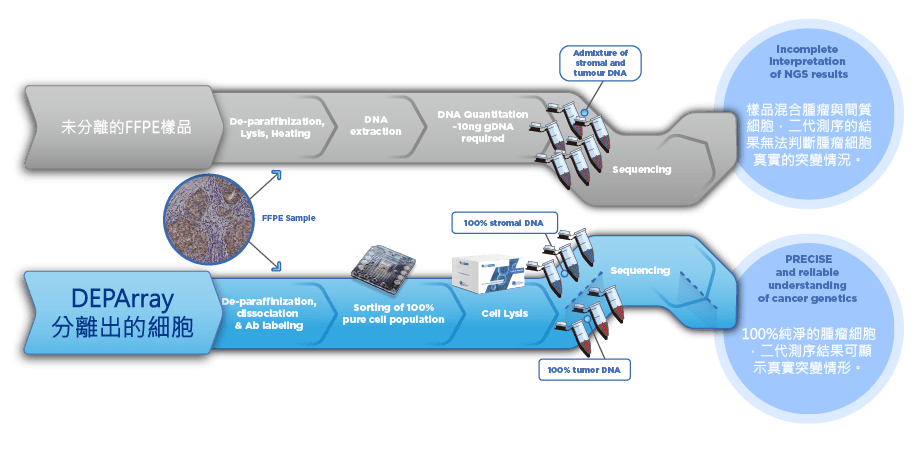
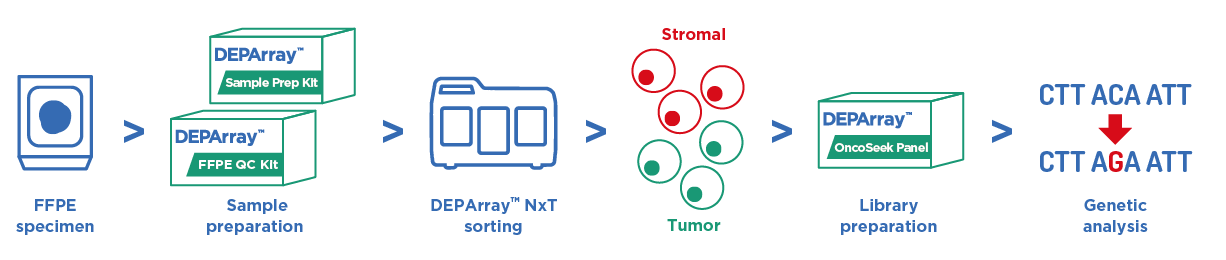
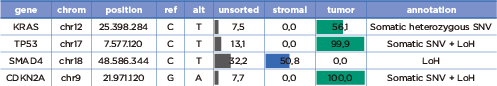
- 組織蠟塊除了使用蘇木精 – 伊紅染色判別外,以 DEPArray™ NxT 分選,可直接準確分選腫瘤細胞 & 基質細胞進行二代測序。
- 精準測得腫瘤序列,有效去除基質細胞干擾
傳統方式 VS DEPArray
完整流程規劃
分辨出腫瘤特異突變基因
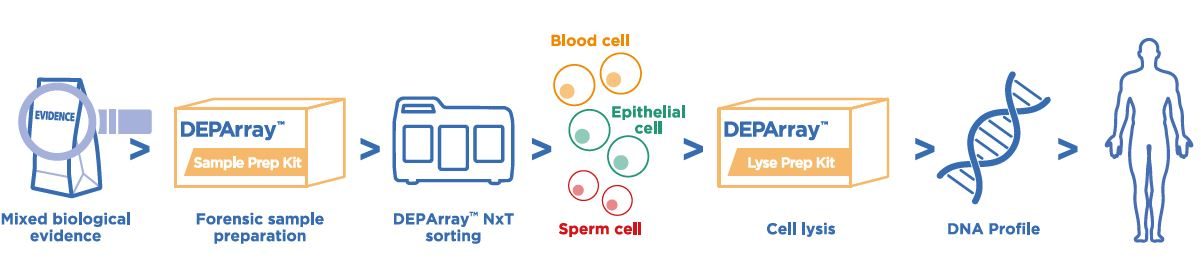
Forensic 法醫鑑定應用
優化檢測程序操作簡易
- 簡易由檢體取得細胞製成的細胞懸浮液。
- 高度自動化且直觀操作模式快速完成實驗。
適合案件檢體
- 僅需 300〜6,000 顆細胞即可上機分選。
- 不同個體細胞比例小於 1:10 的混合生物檢體中,仍可分離出單一來源細胞。
無縫對接常規 STR 分型技術
- 無須更改現有檢測方式,避免耗時的驗證程序。
- 不論案件或數據庫都適用。
獲得單一來源 STR 分型圖譜
- 以全程無污染高純度的單顆細胞或細胞群進行 STR 分型分析。
Forensic 法醫鑑定應用實例
不同類細胞混合檢體的應用實例
2018
Whose blood is it? Application of DEPArray™ technology for the identification of individual/s who contributed blood to a mixed stain
混合 DNA 圖譜的解釋和統計評估通常在法醫 DNA 研究中是常見的特殊挑戰。本研究中將使用 DEPArray™技術分選出的細胞進行 STR 分析結果與早期以傳統方式進行分析的結果做對比。
同類細胞混合檢體的解決方案
2018 Poster
19th European Forensic DNA Working Group Meeting
BLOOD IN BLOOD: isolation and typing of single contributors after DEPArray™ separation
DEPArray™ NxT 使用數位化分選 100%純混合檢體若是混合兩個貢獻者以上的相同種類細胞,只能使用單細胞分析來解決。DEPArray™ NxT 使用數字化分選 100% 純粹的細胞,最多一次可以從一個樣品中回收 48 管單顆細胞。藉由獲得正確的單一 STR 圖譜,可以比對出貢獻者的基因圖譜。的細胞,最多一次可以從一個樣品中回收 48 管單顆細胞。借由獲得正確的單一 STR 圖譜,可以比對出貢獻者的基因圖譜。
2019 Rechtsmedizin
Deconvolution of blood-blood mixtures using DEPArray™ separated single cell STR profiling
應用謀殺案件真實樣本 (刀片上的血斑,保存時間約六週),回收 17 管單顆白細胞,其中 35% 得男性圖譜 (n=6);41% 得女性圖譜 (n=7);24% 無結果 (n=4)。男性圖譜平均完整度為 81%、女性圖譜平均完整度為 79%。整合數個部份圖譜,最終可推導出完整圖譜。
相關產品
完整流程規劃 – 持續優化的流程
循環腫瘤細胞 CTC

組織蠟塊 FFPE

法醫鑑定 Forensic
Ampli1™ LowPass Kit
染色體非整倍體和拷貝數變異 建庫套組
接續 Ampli1™ WGA Kit 產物使用 Ampli1™ LowPass Kit,可輕鬆完成測序前建庫,免除中間複雜的實驗流程,像是純化,去掉 adaptor,再接合等步驟。
DEParray™ 在腫瘤學Oncology的應用
- Mangano et al, Blood Cancer Journal 2019, ” Precise detection of genomic imbalances at single-cell resolution reveals intra-patient heterogeneity in Hodgkin’s lymphoma“
- D’Oronzo et al, Scientific Reports 2019, ” Dissection of major cancer gene variants in subsets of circulating tumor cells in advanced breast cancer“
- Chemi et al, Nature Medicine 2019, ” Pulmonary venous circulating tumor cell dissemination before tumor resection and disease relapse“
- Jang Ho Cho et al, Translational Cancer Research 2019, ” Detection of circulating tumor cells (CTCs) in cerebrospinal fluid of a patient with HER2-overexpressing gastric cancer and single cell analysis of intra-patient heterogeneity of CTCs“
- Brouwer at al, PLOS ONE 2019, ” HER-2 status of circulating tumor cells in a metastatic breast cancer cohort: A comparative study on characterization techniques“
- Pailler et al, Clinical Cancer Research 2019, ” Acquired Resistance Mutations to ALK-Inhibitors Identified by Single Circulating Tumor Cell Sequencing in ALK-Rearranged Non-Small-Cell Lung Cancer“
- Wu et al, Int Journal of Molecular Sciences 2019, ” Nuclear-Biased DUSP6 Expression is Associated with Cancer Spreading Including Brain Metastasis in Triple-Negative Breast Cancer“
- Cariati et al, Int Journal of Molecular Sciences 2019, ” Dissecting Intra-Tumor Heterogeneity by the Analysis of Copy Number Variations in Single Cells: The Neuroblastoma Case Study“
- Meyer et al, Human Pathology 2019 ” Co-expression of cytokeratin and vimentin in colorectal Cancer highlights a subset of tumor buds and an atypical Cancer-associated stroma“
- Hodara et al, Journal od Clinical Oncology Insights 2019, ” Multi-parametric liquid biopsy analysis in metastatic prostate cancer“
- Dorssers et al, British Journal of Cancer 2019, ” Molecular heterogeneity and early metastatic clone selection in testicular germ cell cancer development“
- Rihawi et al, Translational Oncology 2019, ” Amplification as a Potential Mechanism of Primary Resistance to Crizotinib in ALK-Rearranged Non-Small Cell Lung Cancer: A Brief Report“
- Vishnoi et al, Cancer Research 2018, ” Targeting USP7 Identifies a MetastasisCompetent State within Bone Marrow–Resident Melanoma CTCs“
- De Laere et al, Clinical Cancer Research 2018, ” TP53 outperforms other androgen receptor biomarkers to predict abiraterone or enzalutamide outcome in metastatic castration-resistant prostate cancer“
- Chalopin et al, Journal of Bone Oncology, 2018 ” Isolation of circulating tumor cells in a preclinical model of osteosarcoma: effect of chemotherapy“
- Petrossian et al, Oncotarget 2018, ” ERα-mediated cell cycle progression is an important requisite for CDK4/6 inhibitor response in HR+ breast cancer“
- Ji Won Lee et al, Journal of Applied Genetics 2018, “Identification of novel mutations in FFPE lung adenocarcinomas using DEPArray sorting technology and next-generation sequencing”
- Ferrarini A et al, PLOS ONE 2018, “A streamlined workflow for single-cells genome-wide copy-number profiling by low-pass sequencing of LM-PCR whole-genome amplification products”
- Boulding et al, Scientific Reports 2018, ” LSD1 activation promotes inducible EMT programs and modulates the tumour microenvironment in breast cancer“
- Paoletti et al, Cancer Research 2017, ” Comprehensive mutation and copy number profiling in archived circulating breast cancer tumor cells documents heterogeneous resistance mechanisms“
- Boral et al, Nature Communications 2017, “Molecular characterization of breast cancer CTCs associated with brain metastasis”
- Mesquita et al, Molecular Oncology 2017, “Molecular analysis of single circulating tumour cells following long-term storage of clinical samples”
- Paolillo et al, Clinical Cancer Research 2017, ” Detection of Activating Estrogen Receptor Gene (ESR1) Mutations in Single Circulating Tumor Cells“
- Palmirotta et al, Cancer Genomics and Proteomics 2017, “Next-generation Sequencing (NGS) Analysis on Single Circulating Tumor Cells (CTCs) with No Need of Whole-genome Amplification (WGA)”
- Rapp C et al, Acta Neuropathol 2017, ““Identification of T cell target antigens in glioblastoma stem‑like cells using an integrated proteomics‑based approach in patient specimens”
- Bingham C et al, Breast Cancer Res Treat 2017, “Mutational studies on single circulating tumor clles isolated form the blood of inflammatory breast cancer patients”
- Tellez Gabriel M et al, European Journal of Cell Biology 2017, “Analysis of gap junctional intercellular communications using a dielectrophoresis-based microchip”
- De Laere B et al, JTS 2016, “Patients with metastatic hormone receptor-positive breast cancer express PIK3CA oncogene mutational heterogeneity in circulating tumor cells”
- Carter L et al, Nature Medicine 2016, “Molecular analysis of circulating tumor cells identifies distinct copy-number profiles in patients with chemo sensitive and chemo refractory small-cell lung cancer”
- Zhaomei Mu et al, IJMC 2016, “Detection and Characterization of Circulating Tumor Associated Cells in Metastatic Breast Cancer”
- Shaw J et al, Clinical Cancer Research 2016, “Mutation analysis of cell-free DNA and single circulating tumor cells in metastatic breast cancer patients with high CTC counts”
- Millner LM et al, Cancer Research Frontiers 2016, “Comprehensive isolation, identification, and nucleic acid analysis of single breast cancer cells: CTC-isoTECH”
- De Luca F et al, Oncotarget 2016, “Mutational analysis of single circulating tumor cells by next generation sequencing in metastatic breast cancer”
- Bulfoni M et al, Breast Cancer Research 2016, “In patients with metastatic breast cancer the identification of circulating tumor cells in epithelial-to-mesenchymal transition is associated with a poor prognosis”
- Yee S.S et al, Molecular Genetics and Genomic Medicine 2016, “A novel approach for next-generation sequencing of circulating tumor cells”
- Bolognesi C et al, Nature Scientific Reports 2016, “Digital Sorting of Pure Cell Populations Enables Unambiguous Genetic Analysis of Heterogeneous Formalin-Fixed Paraffin-Embedded Tumors by Next Generation Sequencing”
- Rothwell G et al, Molecular Oncology 2015, “Genetic profiling of tumours using both circulating free DNA and circulating tumour cells isolated from the same preserved whole blood sample”
- Vishnoi M et al, Nature Scientific Reports 2015, “The isolation and characterization of CTC subsets related to breast cancer dormancy”
- Salvianti et al, Biomolecular Detection and Quantification 2015, ” Feasibility of a workflow for the molecular characterization of singlecells by next generation sequencing”
- Ewelina Krzywinska et al, EBioMedicine 2015, “Identification of Anti-tumor Cells Carrying Natural Killer (NK) Cell Antigens in Patients with Hematological Cancers”
- Maltoni R et al, Cancer Letters 2015, “Circulating tumor cells in early breast cancer: A connection with vascular invasion”
- Pestrin M et al, Molecular Oncology 2014, “Heterogeneity of PIK3CA mutational status at the single cell level in circulating tumor cells from metastatic breast cancer patients”
- Fernandez S.V. et al, Breast Cancer Research 2014, “TP53 mutations detected in circulating tumor cellspresent in the blood of metastatic triple negative breast cancer patients“
- Carpenter E, et al, Frontiers in Oncology 2014, “Dielectrophoretic capture and genetic analisys of single neuroblastoma tumor cells”
- Hodgkinson C.L.et al, Nature Medicine 2014, “Tumorigenicity and genetic profiling of Circulating Tumor Cells in Small Cell Lung Cancer”
- Peeters DJE, et al. British Journal of Cancer 2013, “Semiautomated isolation and molecular characterization of single or highly purified tumour cells from CellSearch enriched blood samples using dielectrophoretic cell sorting”
- Fabbri F, et al. Cancer Letters 2013, “Detection and recovery of circulating colon cancer cells using a dielectrophoresis-based device: KRAS mutation status in pure CTCs”
DEParray™ 在法醫鑑識Forensic的應用
- Anslinger et al, FSI Genetics Supplement Series 2019, “New strategies in the field of mixture deconvolution single cell STR profiling“
- Anslinger et al, Rechtsmedizin 2019, “Deconvolution of blood-blood mixtures using DEPArrayTM separated single cell STR profiling“
- Anslinger K et al, International Journal of Legal Medicine 2018, “Whose blood is it? Application of DEPArray™ technology for the identification of individual/s who contributed blood to a mixed stain“
- Williamson VR et al, FSI Genetics 2018, “Enhanced DNA Mixture Deconvolution of Sexual Offense Samples Using the DEPArrayTM System“
- Aslinger K et al, Rechtsmedizin 2017, “Application of DEPArray™ technology for the isolation of white blood cells from cell mixtures in chimerism analysis”
- Fontana F et al, FSI Genetics 2017, “Isolation and genetic analysis of pure cells from forensic biological mixtures: The precision of a digital approach”
- Hansson O et al, FSI Genetics 2017, “Characterization of artefacts and drop-in events using STR-validator and single-cell analysis”
DEParray™ 在蠟塊包埋FFPE的應用
- Meyer et al, Human Pathology 2019 “Co-expression of cytokeratin and vimentin in colorectal Cancer highlights a subset of tumor buds and an atypical Cancer-associated stroma“
- Dorssers et al, British Journal of Cancer 2019, “Molecular heterogeneity and early metastatic clone selection in testicular germ cell cancer development“
- Isidori et al, BMC Cancer 2018, “Genomic profiles of primary and metastatic esophageal adenocarcinoma identified via digital sorting of pure cell populations: results from a case report“
- Ji Won Lee et al, Journal of Applied Genetics 2018, “Identification of novel mutations in FFPE lung adenocarcinomas using DEPArray sorting technology and next-generation sequencing”
- Bolognesi C et al, Nature Scientific Reports 2016, “Digital Sorting of Pure Cell Populations Enables Unambiguous Genetic Analysis of Heterogeneous Formalin-Fixed Paraffin-Embedded Tumors by Next Generation Sequencing”
DEParray™ 在分離幹細胞Isolation of Stem Cells的應用
- Silvestris et al, Human Reproduction 2018, “In vitro differentiation of human oocytelike cells from oogonial stem cells: single-cell isolation and molecular characterization”
DEParray™ 在蠟塊包埋FFPE應用的社論
- Kathy Liszewski, GEN 2016, “The Next Next Thing in Sequencing”
- Farideh Bischoff, Nicolò Manaresi and Chiara Bolognesi, GEN 2015, “Isolation of Pure Tumor Cell Populations”
DEParray™技術出版品
以下參考文獻是有關於介電泳技術和移動細胞的原理
- Abonnenc M et al, J Immunol 2013, “Lysis-on-Chip of Single Target Cells following Forced Interaction with CTLs or NK Cells on a Dielectrophoresis-Based Array“
- Abonnenc M et al, Anal. Chem. 2013, “Programmable Interactions of Functionalized Single Bioparticles in a Dielectrophoresis-Based Microarray Chip“
- Gagnon ZR, Electrophoresis 2011, “Cellular dielectrophoresis: applications to the characterization, manipulation, separation and patterning of cells“
- Fabbri E et al, J Appl Polymer Sci 2008, “Levitation and movement of tripalmitin-based cationic lipospheres on a dielectrophoresis-based lab-on-a-chip device“
- Borgatti M et al, Int J Mol Med 2008, “New trends in non-invasive prenatal diagnosis: Applications of dielectrophoresis-based Lab-on-a-chip platforms to the identification and manipulation of rare cells“
- Vulto P et al, J Micromech Microeng 2006, “Selective sample recovery of DEP-separated cells and particles by phaseguide-controlled laminar flow“
- Fuchs AB et al, Lab Chip 2005, “Electronic Sorting and Recovery of Single Live Cells From Microlitre Sized Samples“
- Borgatti M et al, Int J Mol Med 2005, “Separation of White Blood Cells From Erythrocytes on a Dielectrophoresis (DEP) Based ‘Lab-On-A-Chip’ Device“
- Borgatti M et al, International Journal Oncology 2005, “Dielectrophoresis-based ‘Lab-on-a-chip’ devices for programmable binding of microspheres to target cells“
- Abonnenc M et al, NanoBiotechnology 2005, “A dielectrophoretic microchip for controlled cell targeting with functionalized microspheres“
- Medoro G et al, IEEE Sensors J 2003, “A lab-on-a-chip for cell detection and manipulation“
- Altomare L et al, Biotechnol and Bioeng 2003, “Levitation and movement of human tumor cells using a printed circuit board device based on software-controlled dielectrophoresis“
- Gascoyne PR et al, Electrophoresis 2002, “Particle separation by dielectrophoresis“
- Archer S et al, Biochem Biophys Res Commun 1999, “Cell Reactions to Dielectrophoretic Manipulation“



